Abstract
Introduction
Chronic lymphocytic leukemia (CLL) is a highly heterogeneous disease characterized by malignant clonal expansion of mature B lymphocytes. Competitive endogenous RNAs(ceRNAs) such as long noncoding RNAs (lncRNAs) and circular RNAs (circ RNAs) have miRNA response elements (MREs) and can bind to miRNAs to influence mRNA expression. An increasing number of studies have shown that the ceRNA network played an important role in the initiation and progression of tumors. However, the roles and functions of the ceRNA network in chronic lymphocytic leukemia (CLL) are still unclear. This study aims to explore the molecular mechanism of CLL and provide potential prognostic markers and therapeutic targets through the integrated analysis of the ceRNA network in CLL.
Methods
The expression profile of RNAs of CLL patients, CLL cell lines (MEC1 and EHEB) and healthy group were obtained by the illumina sequencing. R software was used for functional enrichment analysis. The data in the genome microarray map GSE22762 was used for survival analysis. The circRNA-miRNA-mRNA ceRNA networks were visualized by Cytoscape 3.7.2. The expression of the circRNA hsa_circ_0007675/hsa-miR-185-3p/TCF7L1 axis were verified by Quantitative real-time PCR and the correlation between hsa_circ_0007675 and TCF7L1 was analyzed.
Results
In total, we identified 57 differentially expressed mRNAs (DEmRNAs), 1391 DElncRNAs, 335 DEmiRNAs and 2413 DEcircRNAs by comparing CLL patients with healthy donors. Meanwhile, 482 mRNAs, 6085 lncRNAs, 302 miRNAs and 1847 circRNAs were explored differently expressed between CLL cell lines and healthy donors.
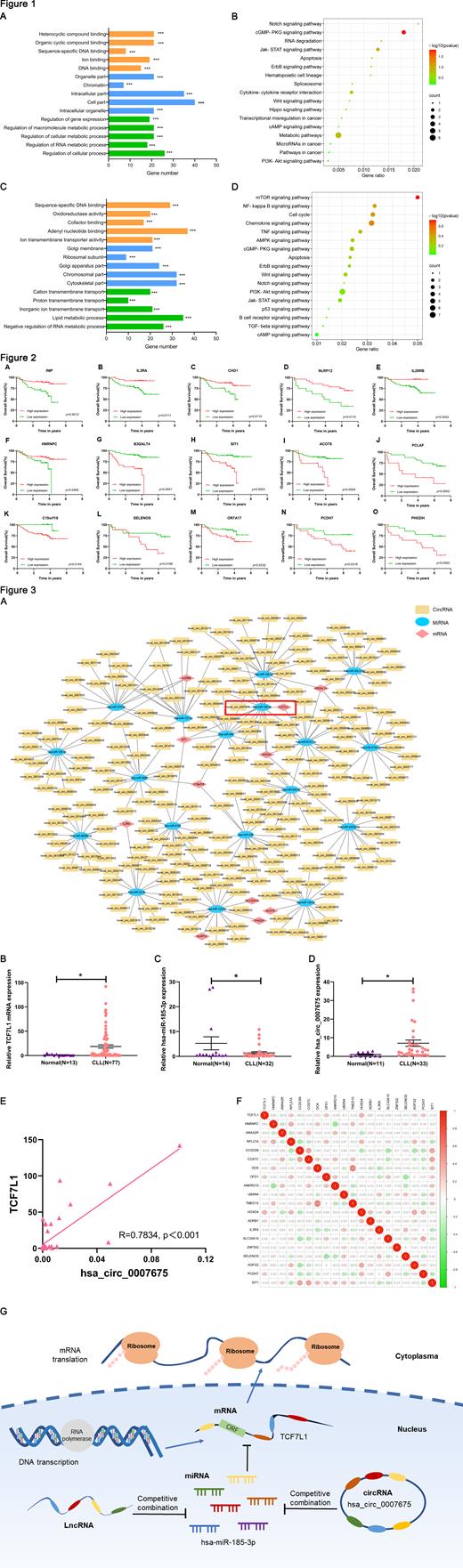
GO analysis results showed that the functions of differentially expressed genes (DEGs) between CLL patients and control are mainly enriched in sequence−specific DNA binding, chromatin and gene expression (Figure 1A) while between CLL cell lines and control they were mainly enriched in oxidoreductase activity, ribosomal subunit and lipid metabolism (Figure 1C). KEGG pathway analysis revealed that the DEGs between CLL patients and control were mainly enriched in Notch signaling pathway, JAK-STAT signaling pathway and cGMP-PKG signaling pathway (Figure 1B). Meanwhile between CLL cell lines and control, DEGs were mainly enriched in mTOR signaling pathway, cell cycle and p53 signaling pathway (Figure 1D).
The survival analyses showed that 15 DEGs (INIP, IL3RA, CHD1, NLRP12, IL20RB, HNRNPC, B3GALT4, SIT1, ACOT8, PCLAF, C19orf18, SELENOS, OR7A17, PCDH7, PHGDH) were significantly differentially expressed in the survival analyses. The overall survival of the high expression group of INIP, IL3RA, CHD1, NLRP12, IL20RB and HNRNPC were higher than that of the low expression group (Figure 2A-F) while the overall survival of the low expression group of B3GALT4, SIT1, ACOT8, PCLAF, C19orf18, SELENOS, OR7A17, PCDH7 and PHGDH were higher than that of the high expression group (Figure 2G-O).
The ceRNA network were built by Cytoscape3.7.2. In total, 11 mRNA nodes, 19 miRNA nodes, 251 circRNA nodes were identified as differentially expressed profiles between CLL patients and control (Figure 3A). We verified the circRNA hsa_circ_0007675/hsa-miR-185-3p/TCF7L1 axis. Compared with normal people, the expression of TCF7L1 and hsa_circ_0007675 in patient specimens were significantly increased (p<0.05; Figure 3B, D) whereas the expression of hsa-miR-185-3p was downregulated (p<0.05; Figure 3C). TCF7L1 and hsa_circ_0007675 were positively correlated (p<0.001, R=0.7834; Figure 3E). The correlation analysis of TCF7L1 and other genes were shown in Figure 3F. The interaction mechanism between them is that hsa_circ_0007675 can sponge hsa-miR-185-3p, thereby inhibiting the inhibitory effect of hsa-miR-185-3p on TCF7L1 and finally upregulate the expression of TCF7L1(Figure 3G).
Conclusions
In this study, we identified the expression profile of RNAs in CLL patients and CLL cell lines. Functional enrichment analysis and survival analysis revealed the potential functions of DEGs. The ceRNA network we established can help to further understand the pathogenesis of CLL and provide potential prognostic biomarkers and novel therapeutic targets.
Keywords: Chronic lymphocytic leukemia; Competing endogenous RNA; Non-coding RNAs; Prognostic biomarkers; Therapeutic targets
No relevant conflicts of interest to declare.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal